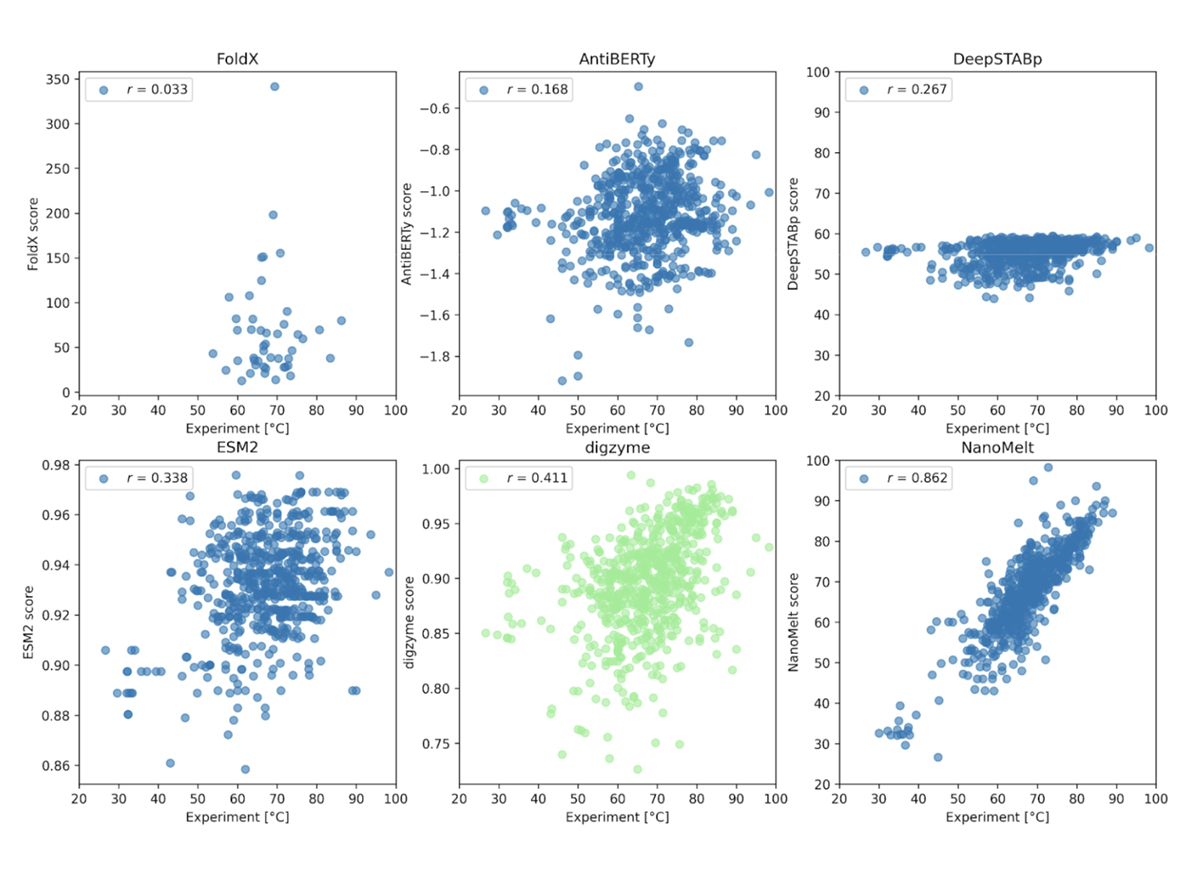
Compare the predictive accuracy of the machine learning model Spotlight™ for enzyme variants’ activity with prior research.
Summary
Our company offers a service called Spotlight that uses a machine learning model to suggest mutants that improve properties such as enzyme activity and thermostability. We input target enzyme sequences into a pre-trained model using various enzymes to predict mutants with improved activity and thermostability for that enzyme. In this tech blog, we have verified the predictive accuracy of Spotlight™ compared to previous research.
The previous research used for comparison.
In Li et al., 2022, a machine learning model (DLKcat) was created to predict kcat using enzyme amino acid sequences and compounds as input information. To ensure equality in the comparison, we utilized the DLKcat machine learning model algorithm and reconstructed the model using the same training data as Spotlight™, namely the kcat entries from BRENDA. We compared the predicted kcat values of the mutants by the reconstructed DLKcat and Spotlight and evaluated which values were closer to the actual measured values. For this study, we extracted entries from BRENDA, ensuring that only wild type (WT) and single mutant variants were included. Our focus was to compare the sensitivity for a single mutation between the two models.
Results
1. Construction of the machine learning model using BRENDA’s kcat (Turnover Number) data.
Entries for variants with reported kcat values were extracted from BRENDA, including entries for the corresponding wild-type (WT) sequences and information about the compounds used to measure kcat. While ensuring no bias toward specific enzyme families, the entries were divided into a 3:1 ratio of training to test data. Training data consisted of 3,969 entries with an increased kcat, 2,985 entries with an unchanged kcat, and 8,296 entries with a decreased kcat (Figure 1). The test data consisted of 792 entries with an increased kcat, 748 entries with an unchanged kcat, and 1,926 entries with a decreased kcat (Figure 2).
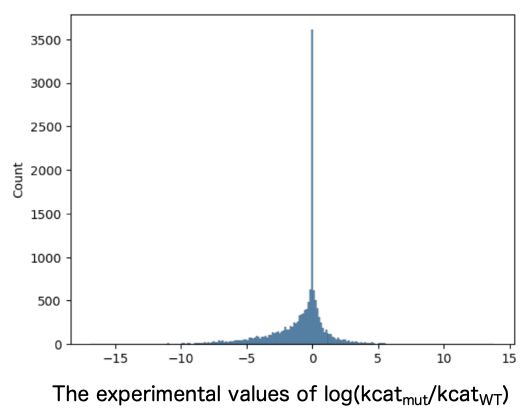
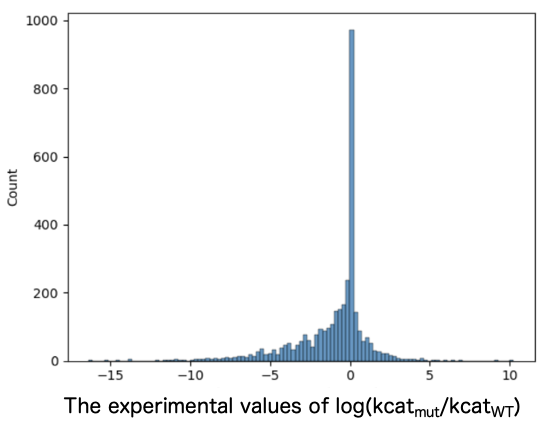
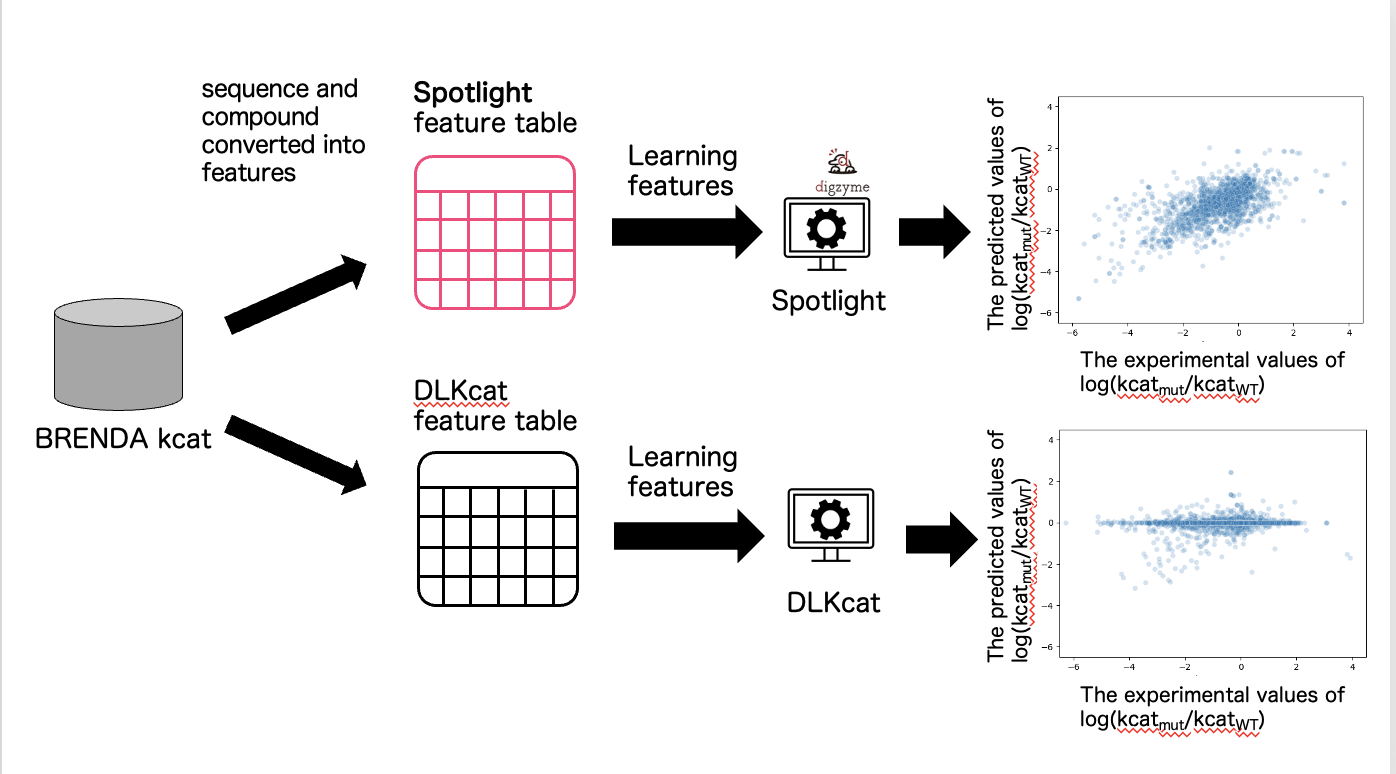
2. Evaluation of mutant/WT ratio of predicted kcat by DLKcat and Spotlight™.
The information from the training data was converted into the format of features required by DLKcat, and a machine learning model was constructed. We converted the training data into the required format of features for Spotlight™ and built a machine learning model (Figure 3).
In the case of DLKcat, the Pearson correlation coefficient between the measured and predicted values of the ratio of the kcat of the mutant to the wild type (WT) kcat was 0.18 (Figure 3). We believe that the reason why the predicted values in DLKcat did not correlate well with the measured values is that DLKcat converts the entire length of the sequence into a vector as a feature, making it difficult for the difference of one amino acid to be reflected in the feature.
In the case of Spotlight™, the Pearson correlation coefficient between the measured and predicted values of the ratio of the kcat of the mutant to the WT kcat was 0.66 (Figure 3). We believe that our Spotlight™ is able to accurately predict the changes caused by single mutations from the WT because it has been devised to reflect the properties of the mutant as a feature.
Conclusion
Our Spotlight™ model was found to more accurately predict changes in activity compared to previous research in cases where only one amino acid mutated.
Acknowledgments
We are grateful for the use of data from the following paper to compare the accuracy of enzyme activity prediction in this study.
Li et al., (2022) Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nature Catalysis.