Exploring the Treasure Trove of Underutilized Sequence Data: Hikoyu Suzuki on Unlocking Value Through Analysis (Employee Interview)
Introduction
This article is a reprint on our tech blog of an interview originally published on our company’s note in November 2024, made available here so that a wider audience can read it. The content reflects the information as of the time of publication.
(*All organization names and job titles mentioned in the article are those at the time of the interview. In the text, we refer to him as “Hikoyu-san” in line with the internal company usage.)
Informatics × Enzyme Development
Hikoyu Suzuki, who studied molecular evolution at Tokyo Institute of Technology and joined digzyme as a big data analytics expert,
shares his insights in this interview on the frontlines of “data-driven enzyme discovery” from the perspective of research and development.

—To start off, could you tell us about how you came to join digzyme?
Even before I seriously started considering a career change, I had registered with JREC-IN, the career support portal provided by the Japan Science and Technology Agency. By chance, I came across a job posting from digzyme there. Since the company originated from my alma mater, Tokyo Institute of Technology, it felt like a meaningful connection. I also felt that my own skills and know-how could be immediately useful, making me a strong candidate—this combination of factors prompted me to apply.
In my previous role, I worked at a company providing analysis services for biological big data—genomes, transcriptomes, and other “omics” datasets. What was similar to digzyme was that many of the projects were close to bespoke, tailor-made assignments.
It’s similar in format to the collaborative research projects I’ve been involved with at digzyme. Of course, it wasn’t specifically focused on enzyme development at that time.
Analyzing big data requires computational expertise, so it’s not something just anyone can do. My work involved handling projects from companies in the food and pharmaceutical sectors, performing analyses tailored to each case, and providing some form of conclusion or insight. You could think of it as being somewhat like consulting work.
—So, in your work, were you combining R&D consulting with biological big data?
Yes, that’s right. I spent about five years at such a venture. A clear example of the projects I handled would be related to antibody therapeutics. Since antibody drugs now occupy the top ranks of pharmaceutical company sales, we often worked on analyses connected to them.
An “antibody” is a protein that can bind specifically to a particular antigen (target). Antibody therapeutics therefore tend to have fewer side effects than conventional drugs and can be highly effective in treating cancers or rare diseases. But antibodies are, of course, proteins, meaning there is a base “human antibody gene.”
The process involves inserting these antibody genes into cultured hamster cells, letting them produce the antibodies in large quantities, purifying them, and turning them into a product. Along the way, you need to consider things like how to optimize the culture methods to produce efficiently at scale, or what sequence to design if you’re developing a new antibody drug.
Nowadays, gaining insights from big data is often part of these considerations. For example, something that works at a small experimental scale doesn’t scale linearly to full production. Scaling up 100 times doesn’t mean you can just produce 100 times as much (laughs). In such cases, we analyze what’s happening by obtaining and studying biological big data, and that’s the kind of support I provided.
—I see. Nakamura-san also mentioned that controlling biological systems at production scale is challenging. What kind of work have you been involved in since joining digzyme?
Right after joining, my work was mainly focused on development based on the concept of synthetic biology, specifically on programs for exploring reaction pathways—including novel reactions—to enable organisms to synthesize target compounds.
For example, if you want to produce a target compound D from a starting compound A, you might need to go through B and C along the way. Because multiple steps are often required, you first need to predict the intermediate reaction pathways; without this, you can’t search for enzymes to catalyze those reactions.
In synthetic biology, when using cultured cells or microbes to produce something useful, a single reaction is rarely sufficient, so predicting the reaction pathway is essential. My role was to explore optimal pathways among many possible options.
Nowadays at digzyme, we mainly handle industrial enzymes, so we don’t need to worry about reaction pathways anymore. We just need to provide the enzymes that meet the customer’s requirements. As a result, my focus has shifted to developing and maintaining libraries of enzyme variants so we can supply
ChatGPT:
—Thank you for touching on the library development! Could you also tell us about your recent work?
I develop and maintain analysis environments that can be used even by WET lab members who aren’t familiar with DRY (computational) analysis. Specifically, this is what’s generally referred to as introducing an IDE (Integrated Development Environment).
For beginners, just installing analysis software can be a major hurdle. Many people might imagine “just download the installer, double-click, and it’s on your computer,” but in bioinformatics, most commonly used tools don’t work that way. From the installation stage, everything is done via the command line—typing commands to instruct the computer—so beginners often get stuck right at this step if they don’t already have some technical knowledge.
To address this, we provide pre-configured environments that remove these obstacles. Introducing an IDE is essentially this solution.
Additionally, many bioinformatics tools don’t have a GUI (Graphical User Interface), which makes them even less approachable. To make them as intuitive as possible, we leverage Jupyter Notebook.
Here’s how it works: WET researchers typically create “lab notebooks” to record experiments. In the same way, we create “analysis notebooks.” These aren’t just simple notes—they contain the executable commands, and even buttons to run those commands. This setup is useful when you want to repeat similar command-line procedures for another project.
In extreme terms, you can perform analyses without fully understanding the underlying programs being used—though of course, it’s not advisable to use it without any understanding at all (laughs).
I work on these environments in collaboration with specialists like Tamura-san (Informatics Specialist, Koichi Tamura) and Isozaki-san (Principal Investigator, Tatsuhiro Isozaki).
—I see. By the way, in what kinds of situations do WET lab members actually perform DRY (computational) analyses?
The main scenario is typically the phase after signing an NDA with a client, when we need to assess feasibility.
Ideally, WET researchers can perform quick analyses during meetings with the client to evaluate feasibility on the spot. This way, even if a DRY team member isn’t present, they can provide immediate, informed feedback, which makes the process very smooth.
Additionally, in the phase of advancing R&D as an actual business project, being able to make decisions about experimental targets and strategies based solely on DRY analysis results—without constant support from computational specialists—offers a significant advantage in terms of development speed.
—Thank you for explaining. What other kinds of work are you involved in?
I’m also involved in handling actual analysis and investigation tasks for individual projects, as well as developing new analysis algorithms. This includes reading the latest research papers and implementing improvements to enhance the accuracy of our existing internal tools. On the biological side, I sometimes work on calculating metrics used for decision-making or assessments.
—What kinds of papers do you usually read?
My background is in molecular biology and molecular evolution, so I’m particularly skilled at analyses involving protein amino acid sequences, as well as “quantitative” information linked to sequences, such as genomes and transcriptomes. As a result, I often read papers that deal directly with gene sequences.
Within the “notebooks” I mentioned earlier, I examine these papers to see if any of the sequence data processing methods can be effectively applied to our analyses.
—While Tamura-san, another informatics specialist, focuses on the structural aspects, Hikoyu-san, you advance analyses daily from the perspective of gene sequences, correct?
That’s right. Our areas of expertise are nicely complementary, so at digzyme we can analyze enzymes from multiple perspectives. I think that’s one of our key strengths.
—Next, could you tell us what aspects of working at digzyme you find most rewarding?
What I find most rewarding is that the results we develop are highly likely to be used directly as a means to create new value for our clients and generate profit for the company.
Many of our clients continue to work with us long after the initial PoC, which serves as a “test of whether we can meet real-world needs.” I can’t share many details due to confidentiality, but I feel that clients recognize the significant synergy they gain from working with digzyme.
Going forward, I want to continue providing new value in response to client needs and make meaningful contributions.
—Hikoyu-san, how do you view the provision of new value that is uniquely possible at digzyme?
As I mentioned earlier, as a premise, there is already a vast amount of genetic information related to enzymes available as big data. Among this data, many sequences have been determined, but their functions are not well understood.
Just because a sequence exists doesn’t mean it lacks function—it’s naturally occurring, and there’s a high likelihood that many of these enzymes can catalyze reactions useful to humanity. How do we find them? Conducting experiments one by one would be impossible. Instead, we first narrow down the search computationally—identifying “this area looks promising”—and of course, experiments are eventually necessary, but this approach maximizes the probability of success with minimal experimental effort. That initial filtering is extremely important, and I believe it’s the essence of digzyme’s technology.
In line with our mission at the time—“Let’s go meet the enzymes that will change the world” (as of November 2024; now updated to “Catalyzing breakthrough options in manufacturing.
Creating lasting vitality for people and Earth.”)—we want to continue uncovering the treasure trove hidden within big data and bring those enzymes to light.
—Thank you for also touching on the essence of digzyme’s technology and your stated mission!
By the way, could you explain the background as to why so many sequences have been recorded in big data but their functions remain poorly understood?
Originally, the practice began with individual researchers registering the sequences of the genes they were studying, so that later, others could verify them or reuse the data in different studies. However, the situation changed drastically as DNA sequencing technology improved.
Especially from the 2010s onward, the widespread adoption of next-generation sequencers made it possible to cheaply decode massive amounts of DNA sequences. Previously, researchers would typically target specific genes using PCR and then sequence them, but now it became feasible to “just read the whole genome!” or “sequence all the RNA being transcribed!”—essentially, a “sequence everything” approach.
As a result, huge volumes of sequence data started accumulating continuously across a wide variety of organisms. While this data has high potential for reuse in other research, the researchers who generated it usually only examine the parts relevant to their own studies.
On the other hand, researchers who have been active for a long time often notice that data collected in other studies can be valuable from the perspective of their own work. This insight led digzyme to adopt the idea of focusing on the reusability of data from a researcher’s point of view. Going forward, we aim to continue uncovering data that holds the potential to address unexplored future needs, mining valuable information that is still hidden.
—I see, that really helps me understand the background. Hikoyu-san, you have such a wide range of expertise, but could you tell us more about what you specialized in at university?
During my student years, I conducted research on evolution under Prof. Norihiro Okada (Emeritus Professor, Tokyo Institute of Technology) and Prof. Masato Nikaido (Associate Professor, Tokyo University of Science, formerly Tokyo Institute of Technology). Specifically, I analyzed molecular-level data such as genes and proteins to investigate evolutionary relationships among different organisms and, from there, explored how genes evolve and the functions of ancestral genes.
At that time, Okada Lab focused on cichlid fishes in the three major lakes of East Africa—Lake Victoria, Lake Malawi, and Lake Tanganyika. These freshwater fishes are highly diverse morphologically and ecologically, and each lake hosts many endemic species, making them a globally recognized model for studying evolution and speciation. The lab energetically pursued research aiming to explain the enormous diversity of cichlids through their genes.
Around the same time, the team led by Prof. Yohei Terai (Associate Professor, SOKENDAI) proposed the sensory drive hypothesis, suggesting that evolution of sensory genes—such as those involved in vision—can promote speciation. Inspired by this, Prof. Nikaido’s team examined other sensory systems, focusing on olfaction.
I specifically focused on olfactory receptor genes, which detect diverse odor molecules and transmit signals to cells. In fish, the olfactory rosette—a petal-like structure visible to the naked eye when the nostrils are opened—houses numerous olfactory neurons that enable fish to sense odors in water. Each of these neurons is believed to express a different olfactory receptor gene. My WET experiments involved classifying these genes phylogenetically, visualizing the neurons expressing each gene, and using olfactory marker proteins to map the neurons expressing these genes.
Over time, my research shifted slightly from cichlids to the evolution of olfactory receptor genes themselves. For instance, Prof. Nikaido had discovered that while most vertebrates possess a single olfactory marker protein gene, teleost fishes have two. I investigated the expression of these two genes in detail and found that they are expressed in different olfactory neurons, and one is also expressed in horizontal cells of the retina—revealing that a gene previously thought to be exclusive to olfactory neurons is also used in retinal neurons.
Additionally, when analyzing the coelacanth genome, I discovered that several olfactory receptor gene lineages previously thought unique to terrestrial vertebrates are also present in “ancient fishes” like coelacanths and polypterids. I also identified a previously unknown type I pheromone receptor gene (ancV1R) conserved across vertebrates, including ancient fishes. The discovery of ancV1R was published in a paper and announced in a press release by Tokyo Tech, and it remains one of my most cherished research achievements.
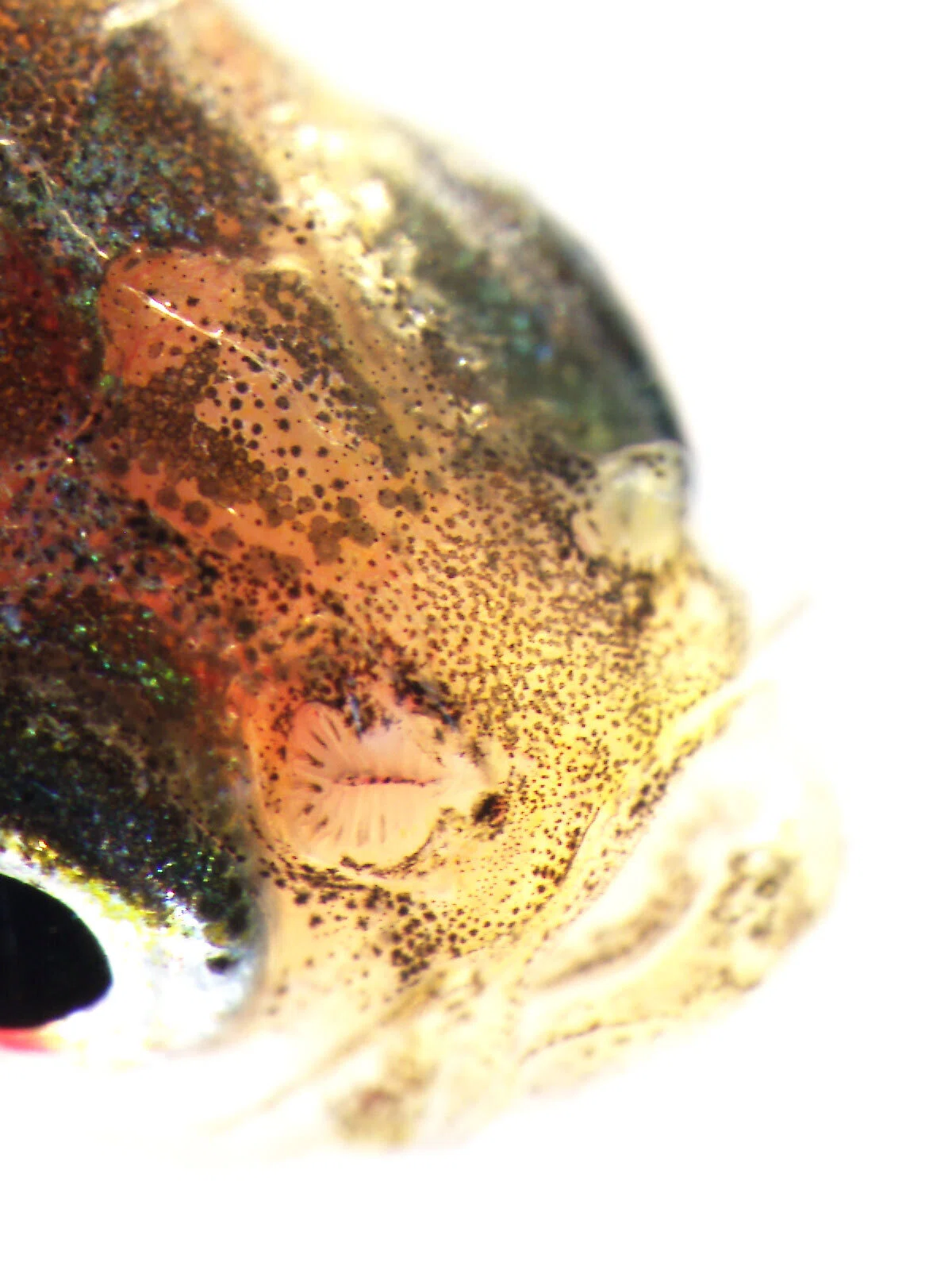
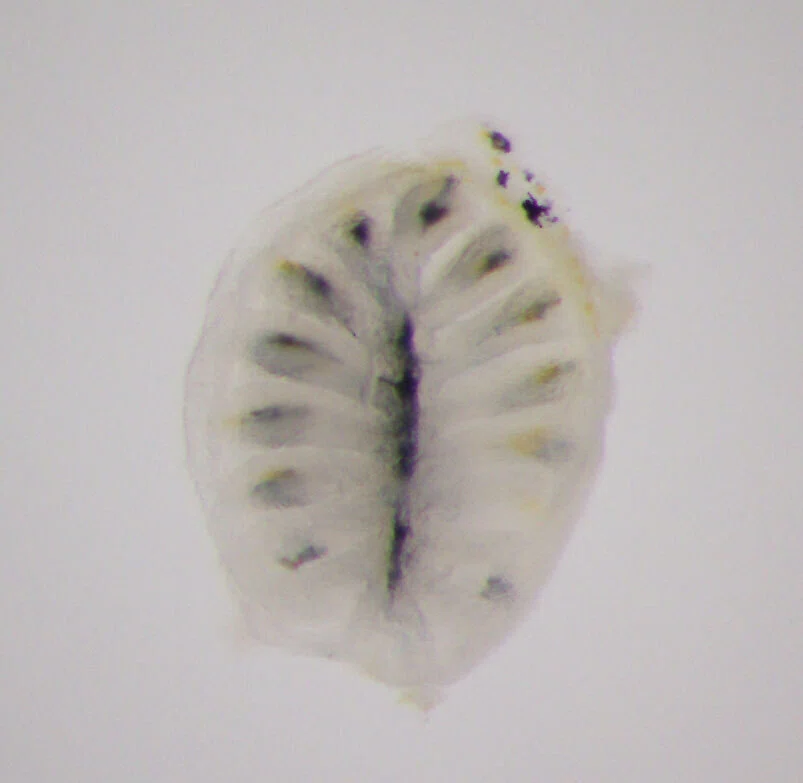
At some point, I also began performing analyses using data generated by next-generation sequencers—devices capable of decoding hundreds of billions to a trillion DNA bases in a short time. There were a few triggers for this shift.
Originally, Okada Lab frequently used analyses of genomes or individual gene sequences. Several assistant professors and postdocs in the lab had self-taught bioinformatics skills and were already applying them to their research, so I thought, “Why not give it a try myself?”
I had some early experience with computer programming as a child, having played around with BASIC, so I started using Perl, a language with similar syntax. Although Perl is less commonly used today, at the time it was widely employed in web application development and excelled at string processing, making it ideal for handling DNA and amino acid sequence data in bioinformatics.
Through this work, I became recognized in the lab as a “student who can do bioinformatics.” Later, Okada Lab’s proposal for a Ministry of Education, Culture, Sports, Science and Technology (MEXT) New Academic Field research project titled “Genetic Basis of Complex Adaptive Trait Evolution” (2010–2015), led by Prof. Mitsuyasu Hasebe (National Institute for Basic Biology), was accepted. Thanks to the recommendations of Prof. Okada and Prof. Nikaido, I was fortunate to participate in the project as a student skilled in bioinformatics.
This project was a challenging endeavor aimed at unraveling aspects of complex evolutionary phenomena across organisms by actively acquiring and utilizing biological big data such as genomes and transcriptomes, leveraging next-generation sequencers, which were just becoming widely adopted at the time.
At the same time, I believe there was an educational aspect, as young researchers like me learned bioinformatics skills, including how to handle next-generation sequencing data. Participating in this project allowed me to gain invaluable experience and skills, which I have been able to apply in both my previous work and my current role at digzyme.
—I see, thank you for explaining that in detail. Hikoyu-san, having been deeply involved in both WET and DRY work, what kind of challenges or new initiatives would you like to pursue at digzyme moving forward?
Exactly—it's about cultivating “dual-skilled” talent in both WET and DRY. When WET researchers gain familiarity with DRY approaches, it can accelerate research, and when DRY researchers understand WET, they can develop solutions that better fit the life sciences environment.
In fact, in my previous position, I was invited as an instructor to give hands-on bioinformatics seminars, explaining techniques to both industry and academic audiences. So at digzyme, I envision something similar—internal seminars or training sessions that help bridge WET and DRY knowledge within the company.
Creating the IDE was also a stepping stone. Of course, simply using the IDE doesn’t automatically build expertise, but moving forward, when new WET members join, we could hold a seminar as a first step—something like, “Let’s try an analysis using the IDE!” I hope to coordinate with CTO Nakamura-san to make this happen.
—Exactly. Because many of digzyme’s DRY engineers come from biological backgrounds, this kind of education is uniquely possible here. In that sense, the composition of our team is indeed a rare and valuable asset.
Exactly. At digzyme, we actually don’t hire people purely from information science backgrounds, because developing enzymes requires deep biological knowledge.
Everyone on our core team—Watari-san, Nakamura-san, Tamura-san, and myself—comes from a biology-related field and then built up our knowledge of information science from that perspective.
Since WET researchers already have a strong background in biology, they have the potential to take the same path—learning DRY techniques from a biological starting point—and digzyme is uniquely positioned to support that kind of training.
Ideally, this type of talent development should be a national policy priority in Japan and addressed across the industry. But in practice, it’s quite challenging for WET researchers to learn DRY skills. The main reason is that opportunities to do so are rare. Unless you’re in an environment like digzyme—where progress depends on understanding both biology and information science—it’s difficult to gain the necessary experience.
—Is there an aspect that makes it highly dependent on the environment?
—There’s also that aspect. In fact, I’ve even heard that in experiment-focused labs, if someone just sits at a computer typing away without doing any wet experiments, they might be seen as “slacking off.”
But in reality, while it’s understandable that people from pure information science backgrounds might avoid studying biology, for biologists, you really can’t separate the two. Now that it’s standard to sequence massive amounts of DNA with next-generation sequencers, avoiding computational approaches puts biologists at a clear disadvantage.
Some might think that the DRY part can just be left to collaborators, but that ends up making the bioinformatics experts extremely busy (laughs). Ideally, even in academia, researchers who study a particular organism should also be able to perform DRY analyses themselves. I believe the core members of the New Academic Fields project I mentioned earlier shared that same ideal, and I think our CEO, Watari-san, would agree as well.
—Finally, do you have a message for future colleagues who are considering applying to digzyme?
—At digzyme, you’ll find an environment where you can work while nurturing both your own curiosity and your desire to contribute to others.
—Thank you very much, Hikoyu-san.
Closing Remarks
▼ Original article is available here (note)
https://note.com/digzyme/n/n7fbee270334b



