Answers to Questions Received at the ifia JAPAN 2025 Exhibition
Introduction
My name is Murase from the Food Business Division.
Our company exhibited at "ifia JAPAN 2025 – The 30th International Food Ingredients & Additives Exhibition and Conference", held at Tokyo Big Sight from Wednesday, May 21 to Friday, May 23, 2025, following our participation last year.
During the exhibition, we had the valuable opportunity to engage directly with many visitors who showed strong interest in our technologies.
At our booth, we introduced our latest initiatives to these attendees. One of the main highlights was the launch of our new solution, “digzyme Custom Enzyme Lab”
(For more details, please refer to our press release:https://prtimes.jp/main/html/rd/p/000000018.000050097.html)


The launch received an overwhelmingly positive response, far exceeding our expectations. Our booth was filled with lively discussions throughout the exhibition, as we received numerous specific questions and inquiries from many visitors each day.
In this special edition of our tech blog, commemorating the launch of “digzyme Custom Enzyme Lab”, we’ve selected some of the most frequently asked questions from the exhibition and provided detailed answers in a Q&A format.
This post is not only for those interested in our new solution, but also for anyone curious about enzyme-based development who may be wondering where to start.
We hope you’ll find useful insights—please read on to the end!
Q: For what types of product development can “digzyme Custom Enzyme Lab” be applied?
A:“digzyme Custom Enzyme Lab” is a flexible solution that can be applied to a wide range of development themes—from specific goals such as improving the efficiency of existing enzyme-based manufacturing processes to broader, more exploratory themes like developing novel food ingredients using enzymes.
By repeatedly exchanging purified enzyme samples and receiving feedback from your in-house evaluations, the development direction can be adjusted flexibly at each stage.
Q: What kind of information is provided with the purified enzyme samples?
A:We perform preliminary testing to confirm enzyme activity and provide a profile including optimal temperature, optimal pH, thermal stability, and pH stability. These data are provided alongside the purified enzyme samples.
Verification in your specific application or evaluation system can be conducted by your team.
Q:What is the quantity of purified enzyme included in the sample?
A:The quantity depends on the development theme and is determined through consultation. As a general guideline, samples are typically provided in volumes of several milliliters of enzyme solution, equivalent to several milligrams of protein.
Q:How do you define or set the initial development timeline?
A:Following a prior evaluation of the requested development theme, we assess the feasibility and propose an initial development timeline.
In most cases, the initial phase—covering in silico enzyme design through to the first delivery of a purified enzyme sample—is completed within 2 to 6 months.
Q:Is non-GMO enzyme development an option?
A:Yes, it is possible. For more details, please refer to the “digzyme Express” introduction page:https://www.digzyme.com/cms/wp-content/uploads/digzyme_Express_ol.pdf
Q:Is “digzyme Custom Enzyme Lab” a solution exclusively for the food industry?
A:“digzyme Custom Enzyme Lab” is a versatile solution available for use not only in the food industry but also in other sectors, including the chemical industry.
Q:If a suitable enzyme is found among the provided purified enzyme samples, what happens next?
A:Enzymes developed via “digzyme Custom Enzyme Lab” can smoothly transition into manufacturing development. digzyme provides comprehensive support throughout the entire process, including manufacturing technology development and regulatory approvals, accompanying you until your project is fully commercialized.
Q:How is intellectual property handled for the developed enzyme library?
A:If you find a promising enzyme among those developed via “digzyme Custom Enzyme Lab” and decide to pursue its commercialization, we are prepared to accommodate your needs flexibly.
This concludes our responses regarding the services provided through “digzyme Custom Enzyme Lab”.
Please feel free to contact us anytime, as we remain flexible and ready to accommodate your specific needs during the actual development process.
Thank you very much for reading through this Q&A.
If you have any questions or require further clarification, please do not hesitate to reach out to us via the contact form below.
[▼ Contact Form]
https://www.digzyme.com/contact/
Search and visualization of polysaccharides containing target monosaccharides from glycan databases.
Summary
My name is Ryu Takayanagi, a second-year master's student at the University of Tokyo, currently interning at digzyme. At university, I have been conducting research related to protein phosphorylation and protein tertiary structures.
In this tech blog, I would like to introduce GlycoSearcher, a new tool we have developed as part of our R&D activities for comprehensive search and visualization of polysaccharides containing target monosaccharides.
In recent years, research and industrial utilization of polysaccharides, such as starch and dietary fiber, have become increasingly active. There is a growing demand for the development of new saccharides, and polysaccharides, in particular, are gaining attention for their high structural diversity. To meet this need, we have developed GlycoSearcher, a tool for comprehensive search of various polysaccharides.
Description formats and databases of polysaccharides
The glyco-compounds we are focusing on have already been reported in numbers exceeding hundreds of thousands and have been databased. To selectively identify polysaccharides that fit specific purposes and apply them in fields such as synthesis pathway exploration and enzyme development, a description format that facilitates computational processing and a comprehensive database are essential.
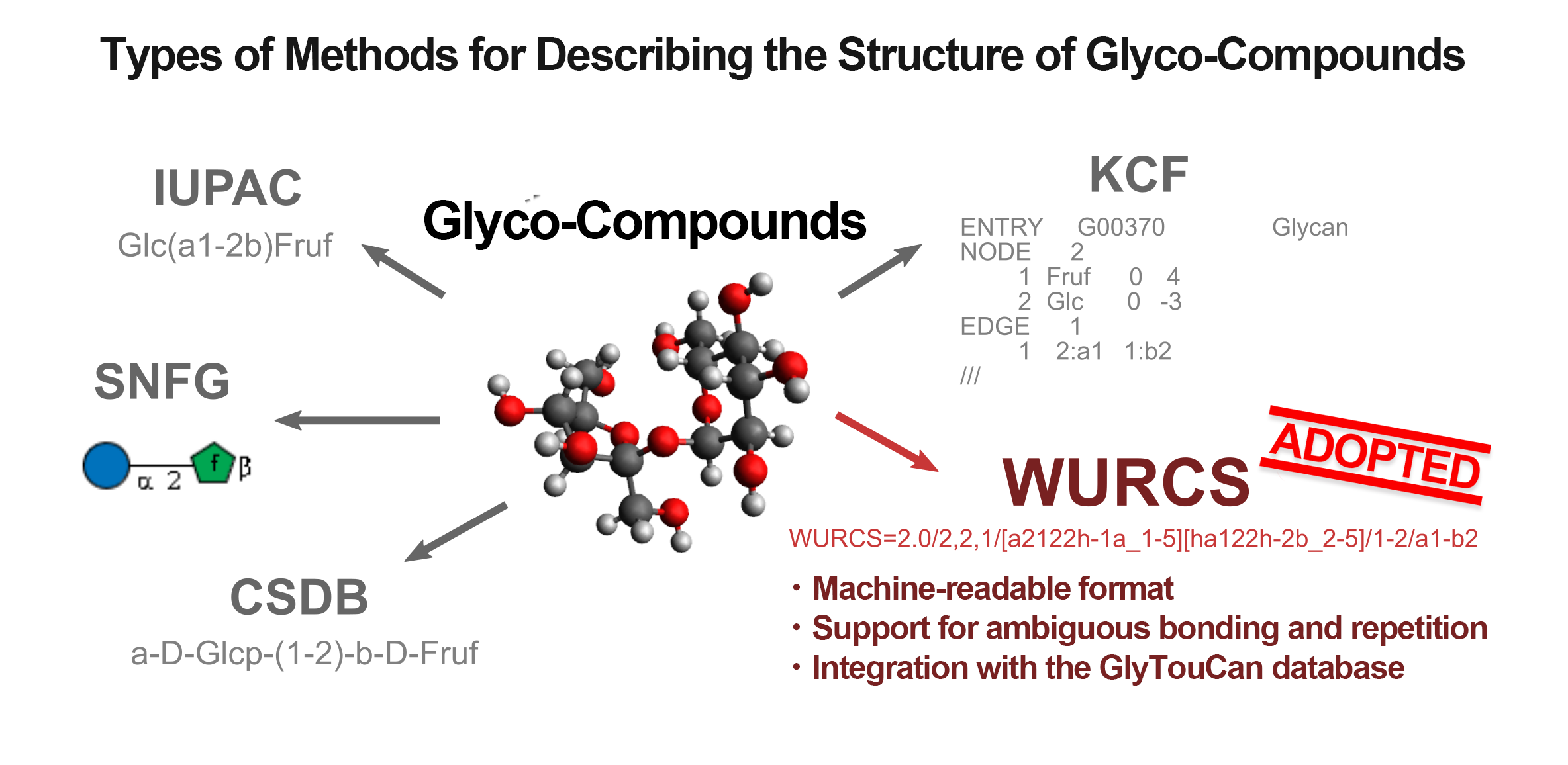
Various methods are known for describing the structure of glyco-compounds (Figure 1). Formats like SNFG and KCF excel in visualization but are not well-suited for advanced computational processing, such as structural information extraction and comparison. On the other hand, the IUPAC format offers a concise structural representation that is readable by both humans and machines, but it struggles with complex and ambiguous expressions, such as repeating units[1]. Therefore, GlycoSearcher employs the WURCS format, which is well-suited for computational processing and can represent repeating units, along with the GlyTouCan database[2], which collects glyco-compound information in the WURCS format.
(Figure 1)
Search Using GlycoSearcher
With GlycoSearcher, it is possible to extract polysaccharides that match specific criteria from a vast number of candidates. For example, you can search for polysaccharides containing particular monosaccharide units such as glucose or galactose. Additionally, it includes a filtering function that allows you to limit the monosaccharide units that make up the polysaccharides. This enables you to list polysaccharides that can be synthesized using a specific monosaccharide as a starting material and other selected sugars.
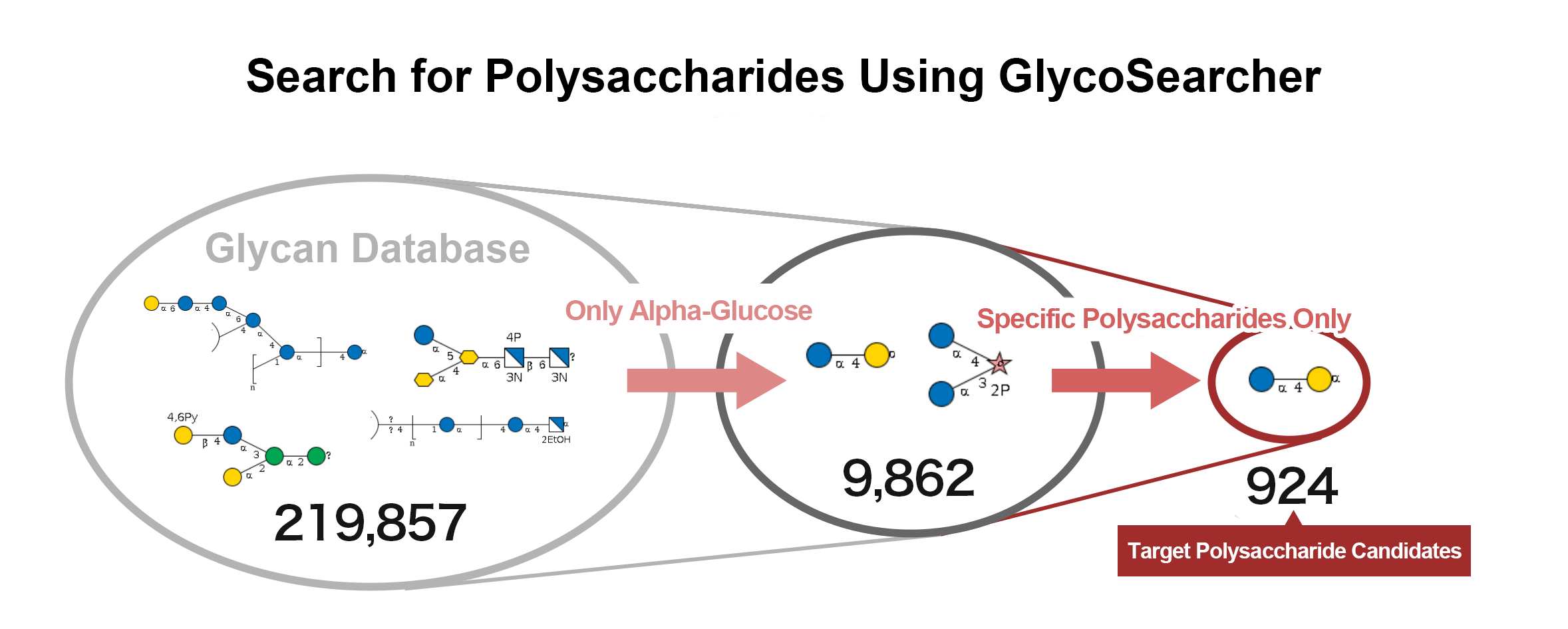
The results of a search for polysaccharides containing α-glucose are shown below (Figure 2). Out of 219,857 glycan structures, 9,862 polysaccharides containing α-glucose were identified. Further narrowing down the search to polysaccharides consisting only of glucose, galactose, and fructose reduced the number of candidates to 924.
(Figure 2)
Visualization and feature extraction of polysaccharide structures
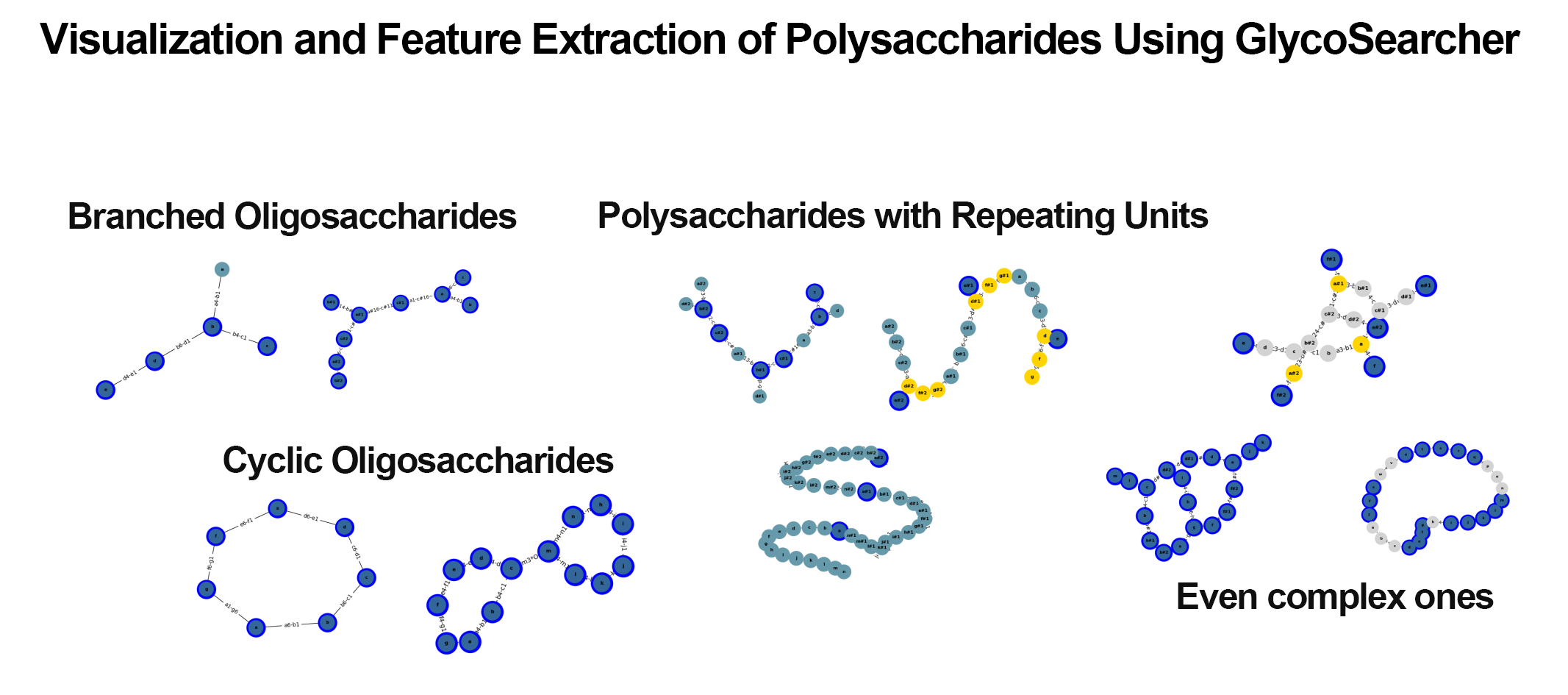
The obtained search results can be effectively visualized and utilized for subsequent applications (Figure 3). By reconstructing polysaccharides in WURCS format as graphs, it is possible to rapidly visualize thousands of search results within minutes. Additionally, for structures with ambiguous repeating units, repeating only a specific number of times allows not only the visualization of the actual structure but also facilitates further computational processing, such as structural comparisons that are challenging when ambiguous.
Since the polysaccharides in the search results are represented as graphs, feature extraction for polysaccharide structures is also possible. For example, computations can determine whether the obtained polysaccharide structures have glucose units at their termini or include specific structures (motifs). Furthermore, by integrating the hit polysaccharides with various databases such as PubChem[3], it is possible to obtain information on their common names and related enzyme information, thus providing insights into reactions involving the polysaccharides.
(Figure 3)
Conclusion
The GlycoSearcher we developed allows for comprehensive searching of target polysaccharides from the database and facilitates further computational processing. Additionally, by extracting information from the identified target polysaccharide candidates and obtaining enzyme information predicted to be involved in their synthesis, we have established a system that links to subsequent enzyme design workflows.
Acknowledgments
The development of GlycoSearcher, including acquiring knowledge about glyco-compounds, was greatly supported by Mr. Isozaki from the Business Development Department. I would like to take this opportunity to express my gratitude.
References
[1] Hosoda, M., & Kinoshita, S. (2021). "Introduction to Glycan-related Informatics." JSBi Bioinformatics Review, 2(1), 87-95.
[2] GlyTouCan. Retrieved from https://glytoucan.org/
[3] PubChem. Retrieved from https://pubchem.ncbi.nlm.nih.gov/
