「digzyme Custom Enzyme Lab」で期待される実用化例:糖鎖構造構築および難分解性物質へのアプローチ
はじめに
2025年5⽉21⽇(水)、22(木)、23 ⽇(金)
と3日間に渡り開催されたifia JAPAN 2025。
昨年と同様、弊社の代表取締役、渡来が出展者プレゼンテーションを行いました。
その様子をYOUTUBEで公開いたしましたので、ぜひご覧ください。
今回の出展者プレゼンテーションでは、2025年5月21日にローンチされた「digzyme Custom Enzyme Lab」についてご紹介しました。当日は、DRY技術(バイオインフォマティクス解析)とWET技術(実験検証)という二つの技術的アプローチに触れつつ、プラットフォーム全体の概要をお伝えしました。
本記事では、その中で取り上げた『「digzyme Custom Enzyme Lab」で期待される実用化例』2件について、渡来の視点を通じて、各事例における技術的ブレイクスルーやin silico設計の裏側をQ&A方式で詳しくご紹介します。
当日のプレゼンテーションでは全体像のご紹介にとどまりましたが、本記事を通じて、「digzyme Custom Enzyme Lab」の実力と可能性をより具体的にご理解いただける内容となっております。ぜひ最後までご覧ください。
まずは一つ目の事例についてです。

「digzyme Custom Enzyme Lab」で期待される実用化例1
Q.この成果の最も大きな意義は何だと考えていますか?
A.糖質は、構成する糖の結合様式の違いによって物性の差が生まれます。in silico技術で、目的の糖鎖構造を作る酵素を狙って探索ができた例は学術的にも稀で、かつ10個という少数の実験で発見できたことは非常に価値が高いと考えています。
Q.これまでのアプローチと比べて、今回のアプローチは何が革新的だったのでしょうか?
A.本件は、結果的には、当時のAlphaFold2に代表される深層学習(DL)ベースの構造予測技術に対して、弊社独自の詳細な分析技術を適用した点が役に立ちました。これまでのhomologyベースのモデルでは、糖鎖構造を作り分ける微妙なタンパク質構造の違いまでは予測することが困難でしたが、当時のAI技術によりそれらの特徴を一定程度捉えられたと考えています。
(なお、現在の生成モデルを用いた最新のAI技術とはギャップがあるため、本稿では便宜的に「AI」とまとめて表現しております。)
Q.チームや関係者のどのような努力がこの成果につながったと思いますか?
A.担当の研究員が、顧客ニーズを詳しく深堀りして当該酵素のスクリーニング基準をうまく設定し、基盤開発メンバーと連携することで解析プログラムを個別に作成し、この成果につながりました。当社では、既存プラットフォームだけで達成できない課題に対しても、フレキシブルにツール開発を行える点が強みだと思っています。
次に、三菱ケミカル株式会社と共同で行った、二つ目の事例についてです。
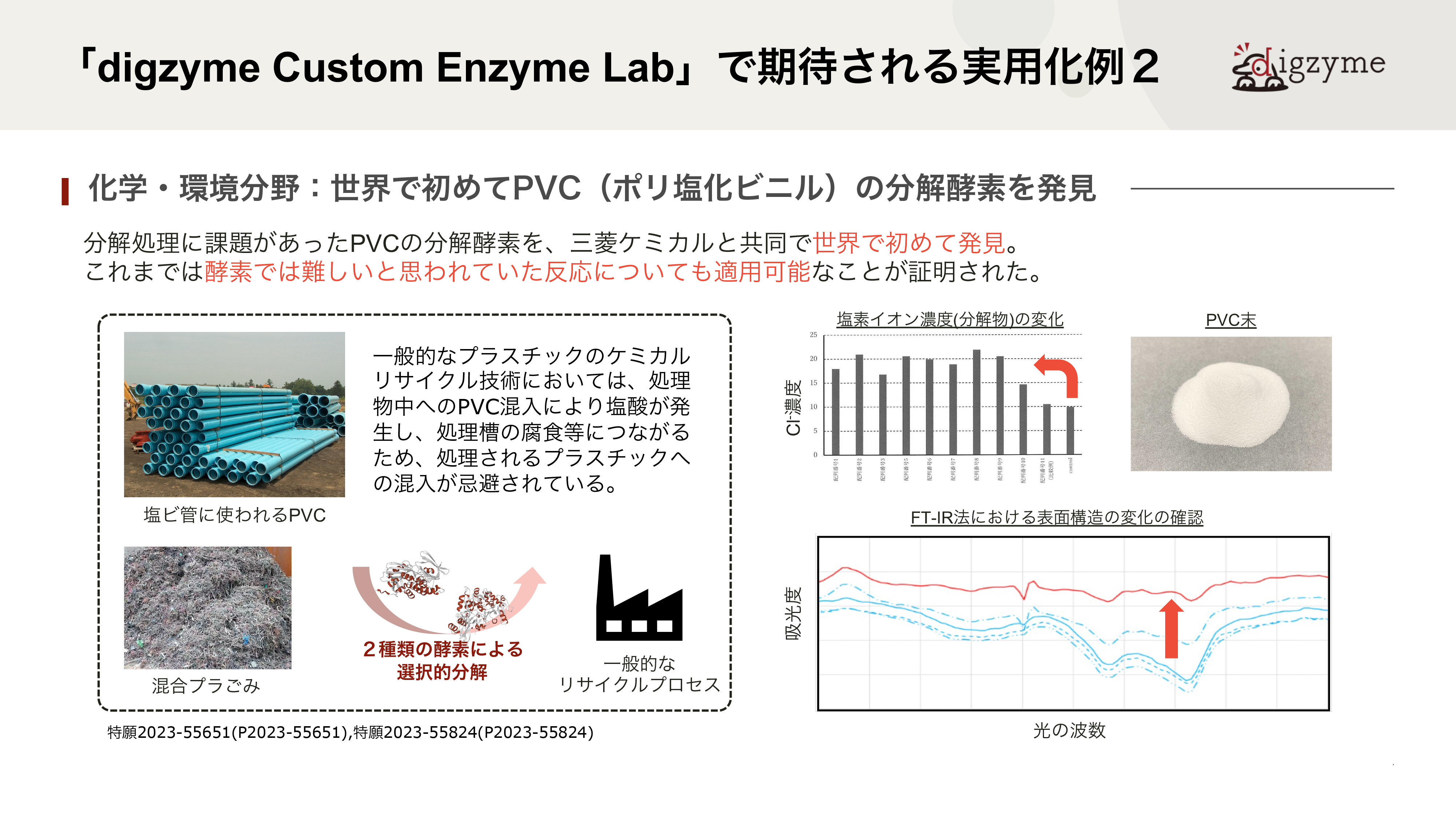
「digzyme Custom Enzyme Lab」で期待される実用化例2
Q.この成果の最も大きな意義は何だと考えていますか?
A.PVCは20世紀から本格的な生産が始まった自然界にない物質で、天然の微生物が進化の過程で分解機構を獲得していないと仮定すると、最適化された酵素は天然からは見つからないはずです。一方で、生物は休眠遺伝子を含めて”最適化されていない”さまざまな遺伝子をゲノムに保有しており、結果として環境変化への適応に活かされるとされています。本件は、その環境適応に寄与しうる酵素をin silicoで人工的に見つける問題とも捉えられるため、難易度が高いテーマでした。
Q.従来、このような酵素を発見するにはどれほどの時間やコストが必要でしたか?
A.近年、人工的なプラスチックの分解酵素を、集積培養に近い形で見出す研究がいくつか見られます。例えば、ある樹脂を海底に一定期間沈めておき、引き上げた後に分解の様子を観察したり、バイオフィルムに含まれる微生物を単離培養したりします。うまくいく場合には、分解酵素を持つ微生物が見つかるので、ゲノム解析やBACライブラリ作成などを通じて分解酵素を同定することが出来ますが、分解が遅いという性質上、どうしても年単位での時間がかかってしまいます。もちろん、分解が観察されず、うまくいかないケースも数多くあると思っています。in silicoでの探索は長くても半年程度で済むため、このような実験時間が長くかかってしまう対象に対してもある程度効果的に用いることが可能です。
終わりに
渡来は、今回の出展者プレゼンテーションを振り返りながら、次のように語っています。
「『digzyme Custom Enzyme Lab』でも、事前の準備期間の中で、これらの共同研究ケースのようなin silicoライブラリを作成して進めていくことができます。高精度のライブラリから精製酵素を試したいお客様におすすめしたいサービスです。」
このコメントが示すように、バイオインフォマティクスを基盤とした酵素設計アプローチは、限られたリソースの中でも実用的な酵素開発を加速しうる可能性を持っています。
今後さらに、酵素の多様な分野への応用が進む中で、「digzyme Custom Enzyme Lab」はその中核を担う技術基盤として重要な役割を果たすと考えられます。



